pyhrf.jde.jde_multi_sess module¶
-
class
pyhrf.jde.jde_multi_sess.BOLDGibbs_Multi_SessSampler(nb_its=3000, obs_hist_pace=-1.0, glob_obs_hist_pace=-1, smpl_hist_pace=-1.0, burnin=0.3, callback=<pyhrf.jde.samplerbase.GSDefaultCallbackHandler object>, response_levels_sess=<pyhrf.jde.jde_multi_sess.NRL_Multi_Sess_Sampler object>, response_levels_mean=<pyhrf.jde.jde_multi_sess.NRLsBar_Drift_Multi_Sess_Sampler object>, beta=<pyhrf.jde.beta.BetaSampler object>, noise_var=<pyhrf.jde.jde_multi_sess.NoiseVariance_Drift_Multi_Sess_Sampler object>, hrf=<pyhrf.jde.jde_multi_sess.HRF_MultiSess_Sampler object>, hrf_var=<pyhrf.jde.hrf.RHSampler object>, mixt_weights=<pyhrf.jde.nrl.bigaussian.MixtureWeightsSampler object>, mixt_params=<pyhrf.jde.nrl.bigaussian.BiGaussMixtureParamsSampler object>, scale=<pyhrf.jde.hrf.ScaleSampler object>, drift=<pyhrf.jde.jde_multi_sess.Drift_MultiSess_Sampler object>, drift_var=<pyhrf.jde.jde_multi_sess.ETASampler_MultiSess object>, stop_crit_threshold=-1, stop_crit_from_start=False, check_final_value=None)¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSampler-
cleanObservables()¶
-
computeFit()¶
-
computePMStimInducedSignal()¶
-
compute_crit_diff(old_vals, means=None)¶
-
default_nb_its= 3000¶
-
finalizeSampling()¶
-
getGlobalOutputs()¶
-
initGlobalObservables()¶
-
inputClass¶ alias of
BOLDSampler_Multi_SessInput
-
parametersComments= {'obs_hist_pace': 'See comment for samplesHistoryPaceSave.', 'smpl_hist_pace': 'To save the samples at each iteration\nIf x<0: no save\n If 0<x<1: define the fraction of iterations for which samples are saved\nIf x>=1: define the step in iterations number between saved samples.\nIf x=1: save samples at each iteration.'}¶
-
parametersToShow= ['nb_its', 'response_levels_sess', 'response_levels_mean', 'hrf', 'hrf_var']¶
-
saveGlobalObservables(it)¶
-
stop_criterion(it)¶
-
updateGlobalObservables()¶
-
-
class
pyhrf.jde.jde_multi_sess.BOLDSampler_Multi_SessInput(data, dt, typeLFD, paramLFD, hrfZc, hrfDuration)¶ Class holding data needed by the sampler : BOLD time courses for each voxel, onsets and voxel topology. It also perform some precalculation such as the convolution matrix based on the onsests (L{stackX}) —- Multi-sessions version
-
buildCosMat(paramLFD, ny)¶
-
buildOtherMatX()¶
-
buildParadigmConvolMatrix(zc, estimDuration, availableDataIndex, parData)¶
-
buildPolyMat(paramLFD, n)¶
-
calcDt(dtMin)¶
-
chewUpOnsets(dt, hrfZc, hrfDuration)¶
-
cleanMem()¶
-
cleanPrecalculations()¶
-
makePrecalculations()¶
-
setLFDMat(paramLFD, typeLFD)¶ Build the low frequency basis from polynomial basis functions.
-
-
class
pyhrf.jde.jde_multi_sess.BiGaussMixtureParams_Multi_Sess_NRLsBar_Sampler(do_sampling=True, use_true_value=False, val_ini=None, hyper_prior_type='Jeffreys', activ_thresh=4.0, var_ci_pr_alpha=2.04, var_ci_pr_beta=0.5, var_ca_pr_alpha=2.01, var_ca_pr_beta=0.5, mean_ca_pr_mean=5.0, mean_ca_pr_var=20.0)¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSamplerVariable-
I_MEAN_CA= 0¶
-
I_VAR_CA= 1¶
-
I_VAR_CI= 2¶
-
L_CA= 1¶
-
L_CI= 0¶
-
NB_PARAMS= 3¶
-
PARAMS_NAMES= ['Mean_Activ', 'Var_Activ', 'Var_Inactiv']¶
-
checkAndSetInitValue(variables)¶
-
computeWithJeffreyPriors(j, cardCIj, cardCAj)¶
-
computeWithProperPriors(j, cardCIj, cardCAj)¶
-
finalizeSampling()¶
-
getCurrentMeans()¶
-
getCurrentVars()¶
-
getOutputs()¶
-
get_string_value(v)¶
-
linkToData(dataInput)¶
-
parametersComments= {'activ_thresh': 'Threshold for the max activ mean above which the region is considered activating', 'hyper_prior_type': "Either 'proper' or 'Jeffreys'"}¶
-
parametersToShow= []¶
-
sampleNextInternal(variables)¶
-
updateObsersables()¶
-
-
class
pyhrf.jde.jde_multi_sess.Drift_MultiSess_Sampler(do_sampling=True, use_true_value=False, val_ini=None)¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSamplerVariable-
checkAndSetInitValue(variables)¶
-
getOutputs()¶
-
get_accuracy(abs_error, rel_error, fv, tv, atol, rtol)¶
-
get_final_value()¶
-
get_true_value()¶
-
linkToData(dataInput)¶
-
sampleNextAlt(variables)¶
-
sampleNextInternal(variables)¶
-
updateNorm()¶
-
-
class
pyhrf.jde.jde_multi_sess.ETASampler_MultiSess(do_sampling=True, use_true_value=False, val_ini=array([ 1.]))¶ Bases:
pyhrf.jde.drift.ETASampler-
linkToData(dataInput)¶
-
sampleNextInternal(variables)¶
-
-
class
pyhrf.jde.jde_multi_sess.HRF_MultiSess_Sampler(do_sampling=True, use_true_value=False, val_ini=None, duration=25.0, zero_constraint=True, normalise=1.0, deriv_order=2, covar_hack=False, prior_type='voxelwiseIID', do_voxelwise_outputs=False, compute_ah_online=False)¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSamplerVariableHRF sampler for multisession model
-
calcXh(hrf)¶
-
checkAndSetInitValue(variables)¶
-
computeStDS_StDY(rb_allSess, nrls_allSess, aa_allSess)¶
-
computeStDS_StDY_from_HRFSampler(rb, nrls, aa)¶ just for comparison purpose. Should be removed in the end.
-
computeStDS_StDY_one_session(rb, nrls, aa, sess)¶
-
finalizeSampling()¶
-
getCurrentVar()¶
-
getFinalVar()¶
-
getOutputs()¶
-
getScaleFactor()¶
-
get_accuracy(abs_error, rel_error, fv, tv, atol, rtol)¶
-
initObservables()¶
-
linkToData(dataInput)¶
-
parametersComments= {'prior_type': 'Type of prior:\n - "singleHRF": one HRF modelled for the whole parcel ~N(0,v_h*R).\n - "voxelwiseIID": one HRF per voxel, all HRFs are iid ~N(0,v_h*R).', 'do_sampling': 'Flag for the HRF estimation (True or False).\nIf set to False then the HRF is fixed to a canonical form.', 'normalise': 'If 1. : Normalise samples of Hrf and NRLs when they are sampled.\nIf 0. : Normalise posterior means of Hrf and NRLs when they are sampled.\nelse : Do not normalise.', 'duration': 'HRF length in seconds', 'covar_hack': 'Divide the term coming from the likelihood by the nb of voxels\n when computing the posterior covariance. The aim is to balance\n the contribution coming from the prior with that coming from the likelihood.\n Note: this hack is only taken into account when "singleHRf" is used for "prior_type"', 'zero_constraint': 'If True: impose first and last value = 0.\nIf False: no constraint.'}¶
-
parametersToShow= ['do_sampling', 'duration', 'zero_constraint']¶
-
reportCurrentVal()¶
-
sampleNextAlt(variables)¶
-
sampleNextInternal(variables)¶
-
samplingWarmUp(variables)¶
-
setFinalValue()¶
-
updateNorm()¶
-
updateObsersables()¶
-
updateXh()¶
-
-
class
pyhrf.jde.jde_multi_sess.NRL_Multi_Sess_Sampler(do_sampling=True, val_ini=None, use_true_value=False)¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSamplerVariable-
checkAndSetInitValue(variables)¶
-
cleanMemory()¶
-
computeAA(nrls, destaa)¶
-
computeComponentsApost(s, m, varXh)¶
-
computeVarYTildeSessionOpt(varXh, s)¶
-
finalizeSampling()¶
-
getOutputs()¶
-
get_accuracy(abs_error, rel_error, fv, tv, atol, rtol)¶
-
is_accurate()¶
-
linkToData(dataInput)¶
-
sampleNextAlt(variables)¶
-
sampleNextInternal(variables)¶
-
samplingWarmUp(variables)¶ #TODO : comment
-
saveCurrentValue(it)¶
-
-
class
pyhrf.jde.jde_multi_sess.NRLsBar_Drift_Multi_Sess_Sampler(do_sampling=True, val_ini=None, contrasts={}, do_label_sampling=True, use_true_nrls=False, use_true_labels=False, labels_ini=None, ppm_proba_threshold=0.05, ppm_value_threshold=0, ppm_value_multi_threshold=array([ 0., 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1., 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2., 2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9, 3., 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9, 4. ]), mean_activation_threshold=4, rescale_results=False, wip_variance_computation=False)¶ Bases:
pyhrf.jde.nrl.bigaussian.NRLSamplerClass handling the Gibbs sampling of Neural Response Levels in the case of joint drift sampling.
-
checkAndSetInitValue(variables)¶
-
get_accuracy(abs_error, rel_error, fv, tv, atol, rtol)¶
-
is_accurate()¶
-
linkToData(dataInput)¶
-
sampleNextAlt(variables)¶
-
sampleNextInternal(variables)¶
-
sampleNrlsSerial(varCI, varCA, meanCA, variables)¶
-
samplingWarmUp(variables)¶ #TODO : comment
-
setFinalValue()¶
-
-
class
pyhrf.jde.jde_multi_sess.NoiseVariance_Drift_Multi_Sess_Sampler(do_sampling=True, use_true_value=False, val_ini=None)¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSamplerVariable-
checkAndSetInitValue(variables)¶
-
linkToData(dataInput)¶
-
sampleNextInternal(variables)¶
-
-
class
pyhrf.jde.jde_multi_sess.Variance_GaussianNRL_Multi_Sess(do_sampling=True, use_true_value=False, val_ini=array([ 1.]))¶ Bases:
pyhrf.xmlio.Initable,pyhrf.jde.samplerbase.GibbsSamplerVariable-
checkAndSetInitValue(variables)¶
-
linkToData(dataInput)¶
-
sampleNextInternal(variables)¶
-
-
pyhrf.jde.jde_multi_sess.b()¶
-
pyhrf.jde.jde_multi_sess.permutation(x)¶ Randomly permute a sequence, or return a permuted range.
If x is a multi-dimensional array, it is only shuffled along its first index.
Parameters: x (int or array_like) – If x is an integer, randomly permute np.arange(x). If x is an array, make a copy and shuffle the elements randomly.Returns: out – Permuted sequence or array range. Return type: ndarray Examples
>>> np.random.permutation(10) array([1, 7, 4, 3, 0, 9, 2, 5, 8, 6])
>>> np.random.permutation([1, 4, 9, 12, 15]) array([15, 1, 9, 4, 12])
>>> arr = np.arange(9).reshape((3, 3)) >>> np.random.permutation(arr) array([[6, 7, 8], [0, 1, 2], [3, 4, 5]])
-
pyhrf.jde.jde_multi_sess.rand(d0, d1, ..., dn)¶ Random values in a given shape.
Create an array of the given shape and populate it with random samples from a uniform distribution over
[0, 1).Parameters: d1, ..., dn (d0,) – The dimensions of the returned array, should all be positive. If no argument is given a single Python float is returned. Returns: out – Random values. Return type: ndarray, shape (d0, d1, ..., dn)See also
random()Notes
This is a convenience function. If you want an interface that takes a shape-tuple as the first argument, refer to np.random.random_sample .
Examples
>>> np.random.rand(3,2) array([[ 0.14022471, 0.96360618], #random [ 0.37601032, 0.25528411], #random [ 0.49313049, 0.94909878]]) #random
-
pyhrf.jde.jde_multi_sess.randn(d0, d1, ..., dn)¶ Return a sample (or samples) from the “standard normal” distribution.
If positive, int_like or int-convertible arguments are provided, randn generates an array of shape
(d0, d1, ..., dn), filled with random floats sampled from a univariate “normal” (Gaussian) distribution of mean 0 and variance 1 (if any of the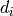 are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.
are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.This is a convenience function. If you want an interface that takes a tuple as the first argument, use numpy.random.standard_normal instead.
Parameters: d1, ..., dn (d0,) – The dimensions of the returned array, should be all positive. If no argument is given a single Python float is returned. Returns: Z – A (d0, d1, ..., dn)-shaped array of floating-point samples from the standard normal distribution, or a single such float if no parameters were supplied.Return type: ndarray or float See also
random.standard_normal()- Similar, but takes a tuple as its argument.
Notes
For random samples from
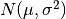 , use:
, use:sigma * np.random.randn(...) + muExamples
>>> np.random.randn() 2.1923875335537315 #random
Two-by-four array of samples from N(3, 6.25):
>>> 2.5 * np.random.randn(2, 4) + 3 array([[-4.49401501, 4.00950034, -1.81814867, 7.29718677], #random [ 0.39924804, 4.68456316, 4.99394529, 4.84057254]]) #random
-
pyhrf.jde.jde_multi_sess.sampleHRF_single_hrf(stLambdaS, stLambdaY, varR, rh, nbColX, nbVox)¶
-
pyhrf.jde.jde_multi_sess.sampleHRF_single_hrf_hack(stLambdaS, stLambdaY, varR, rh, nbColX, nbVox)¶
-
pyhrf.jde.jde_multi_sess.sampleHRF_voxelwise_iid(stLambdaS, stLambdaY, varR, rh, nbColX, nbVox, nbSess)¶
-
pyhrf.jde.jde_multi_sess.simulate_sessions(output_dir, snr_scenario='high_snr', spatial_size='tiny')¶
-
pyhrf.jde.jde_multi_sess.simulate_single_session(output_dir, var_sessions_nrls, cdefs, nrls_bar, labels, labels_vol, v_noise, drift_coeff_var, drift_amplitude)¶