pyhrf.jde.nrl.bigaussian_drift module¶
-
class
pyhrf.jde.nrl.bigaussian_drift.BiGaussMixtureParams_Multi_Sess_NRLsBar_Sampler(parameters=None, xmlHandler=None, xmlLabel=None, xmlComment=None)¶ Bases:
pyhrf.jde.samplerbase.GibbsSamplerVariable#TODO : comment
-
I_MEAN_CA= 0¶
-
I_VAR_CA= 1¶
-
I_VAR_CI= 2¶
-
L_CA= 1¶
-
L_CI= 0¶
-
NB_PARAMS= 3¶
-
PARAMS_NAMES= ['Mean_Activ', 'Var_Activ', 'Var_Inactiv']¶
-
P_ACTIV_THRESH= 'mean_activation_threshold'¶
-
P_HYPER_PRIOR= 'hyperPriorType'¶
-
P_MEAN_CA_PR_MEAN= 'meanCAPrMean'¶
-
P_MEAN_CA_PR_VAR= 'meanCAPrVar'¶
-
P_SAMPLE_FLAG= 'sampleFlag'¶
-
P_USE_TRUE_VALUE= 'useTrueValue'¶
-
P_VAL_INI= 'initialValue'¶
-
P_VAR_CA_PR_ALPHA= 'varCAPrAlpha'¶
-
P_VAR_CA_PR_BETA= 'varCAPrBeta'¶
-
P_VAR_CI_PR_ALPHA= 'varCIPrAlpha'¶
-
P_VAR_CI_PR_BETA= 'varCIPrBeta'¶
-
checkAndSetInitValue(variables)¶
-
computeWithJeffreyPriors(j, cardCIj, cardCAj)¶
-
computeWithProperPriors(j, cardCIj, cardCAj)¶
-
defaultParameters= {'initialValue': None, 'mean_activation_threshold': 4.0, 'varCAPrBeta': 0.5, 'varCIPrAlpha': 2.04, 'meanCAPrVar': 20.0, 'hyperPriorType': 'Jeffrey', 'varCIPrBeta': 2.08, 'sampleFlag': True, 'varCAPrAlpha': 2.01, 'useTrueValue': False, 'meanCAPrMean': 5.0}¶
-
finalizeSampling()¶
-
getCurrentMeans()¶
-
getCurrentVars()¶
-
getOutputs()¶
-
get_string_value(v)¶
-
linkToData(dataInput)¶
-
parametersComments= {'hyperPriorType': "Either 'proper' or 'Jeffrey'", 'mean_activation_threshold': 'Threshold for the max activ mean above which the region is considered activating'}¶
-
parametersToShow= ['initialValue', 'sampleFlag', 'mean_activation_threshold', 'useTrueValue', 'hyperPriorType', 'meanCAPrMean', 'meanCAPrVar', 'varCIPrAlpha', 'varCIPrBeta', 'varCAPrAlpha', 'varCAPrBeta']¶
-
sampleNextInternal(variables)¶
-
updateObsersables()¶
-
-
class
pyhrf.jde.nrl.bigaussian_drift.NRL_Drift_Sampler(do_sampling=True, val_ini=None, contrasts={}, do_label_sampling=True, use_true_nrls=False, use_true_labels=False, labels_ini=None, ppm_proba_threshold=0.05, ppm_value_threshold=0, ppm_value_multi_threshold=array([ 0., 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1., 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2., 2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9, 3., 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9, 4. ]), mean_activation_threshold=4, rescale_results=False, wip_variance_computation=False)¶ Bases:
pyhrf.jde.nrl.bigaussian.NRLSamplerClass handling the Gibbs sampling of Neural Response Levels in the case of joint drift sampling.
-
computeVarYTildeOpt(varXh)¶
-
sampleNextInternal(variables)¶
-
sampleNrlsSerial(rb, h, varCI, varCA, meanCA, gTg, variables)¶
-
-
class
pyhrf.jde.nrl.bigaussian_drift.NRL_Drift_SamplerWithRelVar(do_sampling=True, val_ini=None, contrasts={}, do_label_sampling=True, use_true_nrls=False, use_true_labels=False, labels_ini=None, ppm_proba_threshold=0.05, ppm_value_threshold=0, ppm_value_multi_threshold=array([ 0., 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1., 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2., 2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9, 3., 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9, 4. ]), mean_activation_threshold=4, rescale_results=False, wip_variance_computation=False)¶ Bases:
pyhrf.jde.nrl.bigaussian.NRLSamplerWithRelVarClass handling the Gibbs sampling of Neural Response Levels in the case of joint drift sampling and relevant variable.
-
computeVarYTildeOptWithRelVar(varXh, w)¶
-
sampleNextInternal(variables)¶
-
sampleNrlsSerialWithRelVar(rb, h, gTg, variables, w, t1, t2)¶
-
-
class
pyhrf.jde.nrl.bigaussian_drift.NRLsBar_Drift_Multi_Sess_Sampler(do_sampling=True, val_ini=None, contrasts={}, do_label_sampling=True, use_true_nrls=False, use_true_labels=False, labels_ini=None, ppm_proba_threshold=0.05, ppm_value_threshold=0, ppm_value_multi_threshold=array([ 0., 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1., 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2., 2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9, 3., 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9, 4. ]), mean_activation_threshold=4, rescale_results=False, wip_variance_computation=False)¶ Bases:
pyhrf.jde.nrl.bigaussian.NRLSamplerClass handling the Gibbs sampling of Neural Response Levels in the case of joint drift sampling.
-
checkAndSetInitValue(variables)¶
-
linkToData(dataInput)¶
-
sampleNextAlt(variables)¶
-
sampleNextInternal(variables)¶
-
sampleNrlsSerial(varCI, varCA, meanCA, variables)¶
-
samplingWarmUp(variables)¶ #TODO : comment
-
-
pyhrf.jde.nrl.bigaussian_drift.permutation(x)¶ Randomly permute a sequence, or return a permuted range.
If x is a multi-dimensional array, it is only shuffled along its first index.
Parameters: x (int or array_like) – If x is an integer, randomly permute np.arange(x). If x is an array, make a copy and shuffle the elements randomly.Returns: out – Permuted sequence or array range. Return type: ndarray Examples
>>> np.random.permutation(10) array([1, 7, 4, 3, 0, 9, 2, 5, 8, 6])
>>> np.random.permutation([1, 4, 9, 12, 15]) array([15, 1, 9, 4, 12])
>>> arr = np.arange(9).reshape((3, 3)) >>> np.random.permutation(arr) array([[6, 7, 8], [0, 1, 2], [3, 4, 5]])
-
pyhrf.jde.nrl.bigaussian_drift.rand(d0, d1, ..., dn)¶ Random values in a given shape.
Create an array of the given shape and populate it with random samples from a uniform distribution over
[0, 1).Parameters: d1, ..., dn (d0,) – The dimensions of the returned array, should all be positive. If no argument is given a single Python float is returned. Returns: out – Random values. Return type: ndarray, shape (d0, d1, ..., dn)See also
random()Notes
This is a convenience function. If you want an interface that takes a shape-tuple as the first argument, refer to np.random.random_sample .
Examples
>>> np.random.rand(3,2) array([[ 0.14022471, 0.96360618], #random [ 0.37601032, 0.25528411], #random [ 0.49313049, 0.94909878]]) #random
-
pyhrf.jde.nrl.bigaussian_drift.randn(d0, d1, ..., dn)¶ Return a sample (or samples) from the “standard normal” distribution.
If positive, int_like or int-convertible arguments are provided, randn generates an array of shape
(d0, d1, ..., dn), filled with random floats sampled from a univariate “normal” (Gaussian) distribution of mean 0 and variance 1 (if any of the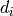 are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.
are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.This is a convenience function. If you want an interface that takes a tuple as the first argument, use numpy.random.standard_normal instead.
Parameters: d1, ..., dn (d0,) – The dimensions of the returned array, should be all positive. If no argument is given a single Python float is returned. Returns: Z – A (d0, d1, ..., dn)-shaped array of floating-point samples from the standard normal distribution, or a single such float if no parameters were supplied.Return type: ndarray or float See also
random.standard_normal()- Similar, but takes a tuple as its argument.
Notes
For random samples from
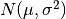 , use:
, use:sigma * np.random.randn(...) + muExamples
>>> np.random.randn() 2.1923875335537315 #random
Two-by-four array of samples from N(3, 6.25):
>>> 2.5 * np.random.randn(2, 4) + 3 array([[-4.49401501, 4.00950034, -1.81814867, 7.29718677], #random [ 0.39924804, 4.68456316, 4.99394529, 4.84057254]]) #random